
"Saint Louis. Born, raised, pre-school through PhD."

"Saint Louis. Born, raised, pre-school through PhD."
I am a research scientist at Meta Reality Labs, working in the area of quantified wearability. My interests lie in parametric face modeling and 3D reconstruction. Previously, I was a PhD candidate with the Department of Computer Science and Engineering at Washington University in St. Louis, advised by Dr. Tao Ju. My research integrated geometry processing, topology, and computer graphics to address challenges in geometry modeling and biomedical and plant imaging. I was being funded by the Imaging Sciences Pathway Fellowship from Washington University in St. Louis. Before enrolling in my PhD program, I earned my bachelor's degree in Computer Science from Washington University in St. Louis in 2017. I graduated in May 2022.
During my PhD, the first area of my work focused on topological optimization for 3D surface reconstruction. I had a paper accepted for publication to SIGGRAPH Asia 2020 about a global optimization algorithm for topological simplification of 3D shapes. Additionally, I am collaborating with Saint Louis University and biologists at the Donald Danforth Plant Science Center to phenotype plants such as corn and sorghum from 3D imaging. My latest work is TopoRoot, a software pipeline for extracting hierarchy and fine-grained traits of Maize roots from X-ray CT images. Additionally, together with Mao Li, Mon-Ray Shao, Christopher N. Topp, and Toby Kellogg, we published a paper which made the cover of New Phytologist Journal. During the summer of 2020 I did a research internship with Facebook Reality Labs.
We present a method for removing unwanted topological features (e.g., islands, handles, cavities) from a sequence of shapes where each shape is nested in the next. Such sequences can be found in nature, such as a multi-layered material or a growing plant root. Existing topology simplification methods are designed for single shapes, and applying them independently to shapes in a sequence may lose the nesting property. We formulate the nesting-constrained simplification task as an optimal labelling problem on a set of candidate shape deletions (cuts) and additions (fills). We explored several optimization strategies, including a greedy heuristic that sequentially propagates labels, a state-space search algorithm that is provably optimal, and a beam-search variant with controllable complexity. Evaluation on synthetic and real-world data shows that our method is as effective as single-shape simplification methods in reducing topological complexity and minimizing geometric changes, and it additionally ensures nesting. Also, the beam-search strategy is found to strike the best balance between optimality and efficiency.
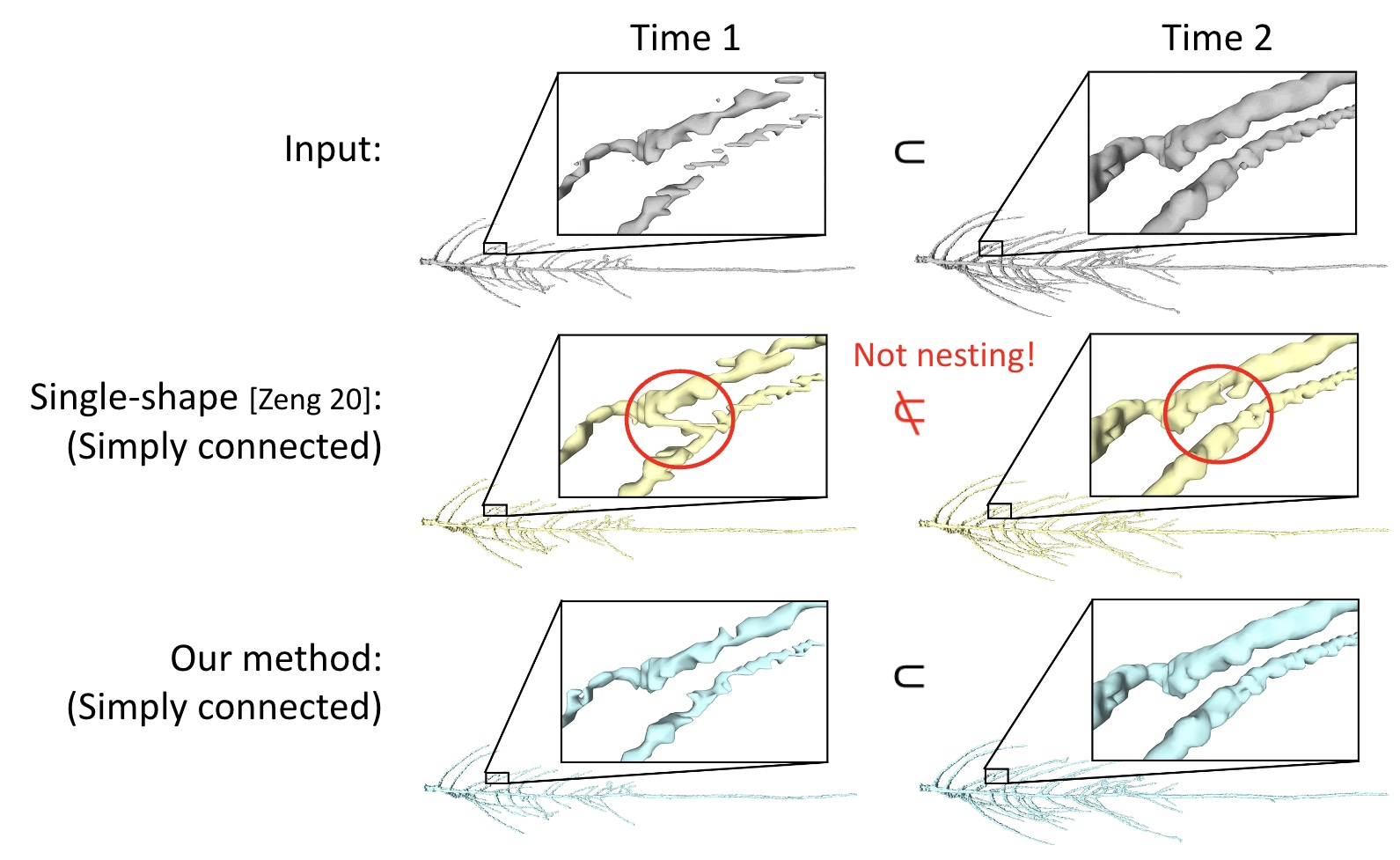
Simplifying the topology of a growing sequence of pen- nycress root (top row, showing two time points) by applying the simplification method [ZCLJ20] to each shape independently (mid- dle row) and by our proposed method (bottom row). While both methods fully simplify the topology of each shape, the results of [ZCLJ20] are no longer nested (e.g., regions highlighted in blue circles) but ours are.
3D imaging, such as X-ray CT and MRI, has been widely deployed to study plant root structures. Many computational tools exist to extract coarse-grained features from 3D root images, such as total volume, root number and total root length. However, methods that can accurately and efficiently compute fine-grained root traits, such as root number and geometry at each hierarchy level, are still lacking. These traits would allow biologists to gain deeper insights into the root system architecture (RSA). We present TopoRoot, a high-throughput computational method that computes fine-grained architectural traits from 3D X-ray CT images of field-excavated maize root crowns. These traits include the number, length, thickness, angle, tortuosity, and number of children for the roots at each level of the hierarchy. TopoRoot combines state-of-the-art algorithms in computer graphics, such as topological simplification and geometric skeletonization, with customized heuristics for robustly obtaining the branching structure and hierarchical information. TopoRoot is validated on both real and simulated root images, and in both cases it was shown to improve the accuracy of traits over existing methods. We also demonstrate TopoRoot in differentiating a maize root mutant from its wild type segregant using fine-grained traits. TopoRoot runs within a few minutes on a desktop workstation for volumes at the resolution range of 400^3, without need for human intervention. TopoRoot's improved accuracy and automation make it useful for phenomic studies aimed at finding the genetic basis behind root system architecture and the subsequent development of more productive crops.
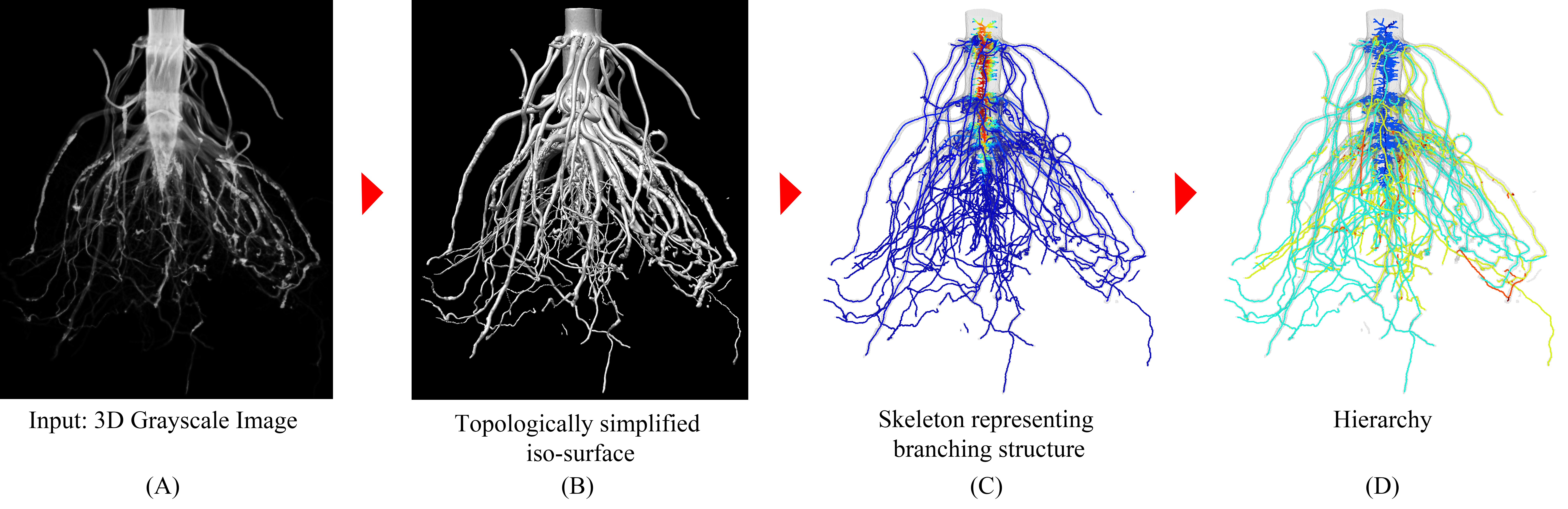
The pipeline of TopoRoot for computing fine-grained traits from a 3D X-ray image. Beginning from a 3D grayscale image (A), TopoRoot first simplifies the topological complexity of the iso-surface (B), then it creates a geometric skeleton capturing the branching structure (C), from which a hierarchy is obtained (D) and the traits are subsequently computed.
We present a novel algorithm for simplifying the topology of a 3D shape, which is characterized by the number of connected components, handles, and cavities. Existing methods either limit their modifications to be only cutting or only filling, or take a heuristic approach to decide where to cut or fill. We consider the problem of finding a globally optimal set of cuts and fills that achieve the simplest topology while minimizing geometric changes. We show that the problem can be formulated as graph labelling, and we solve it by a transformation to the Node-Weighted Steiner Tree problem. When tested on examples with varying levels of topological complexity, the algorithm shows notable improvement over existing simplification methods in both topological simplicity and geometric distortions.
To simplify the topology of a 3D shape (a), performing cutting alone (b) or filling alone (c) results in excessive changes, such as removing large components (box C in (b)), creating long bridges to distant islands (box A in (c)) and large patches to fill in a handle (box B in (c)). Given a set of pre-computed cuts and fills, our method optimally selects a subset of them to maximally simplify topology while minimizing the impact on the geometry (d). (β: number of connected components, handles, and cavities; д: geometric cost)
Inflorescence architecture in plants is often complex and challenging to quantify, particularly for inflorescences of cereal grasses. Methods for capturing inflorescence architecture and for analyzing the resulting data are limited to a few easily captured parameters that may miss the rich underlying diversity. Here, we apply X-ray computed tomography combined with detailed morphometrics, offering new imaging and computational tools to analyze three-dimensional inflorescence architecture. To show the power of this approach, we focus on the panicles of Sorghum bicolor, which vary extensively in numbers, lengths, and angles of primary branches, as well as the three-dimensional shape, size, and distribution of the seed.
In 2018-2019, I organized a dragon dance team with members from around the Saint Louis area, many coming from Washington University and the Saint Louis Modern Chinese School. We called ourselves the Saint Dragon Team and performed at various festivals and holidays including the Chinese Culture Days at the Missouri Botanical Garden, the Gateway Dragon Boat Festival, and the Independence Day and Thanksgiving Day parades in Downtown St. Louis. Shown in this video was our performance for the opening ceremony of the Gateway Dragon Boat Festival in 2018 at Creve Coeur Lake.
I really enjoyed my internship with the Donald Danforth Plant Science center in the summer of 2018 because we got to spend some time outdoors and I got to interact with the subject matter of my research. The first two pictures show me planting Sorghum seeds during a field trip to some farmlands near Columbia, MO at the beginning of the internship, while the last one shows me washing the dirt off of their roots after harvesting.


